-Search query
-Search result
Showing all 45 items for (author: dong & yy)

EMDB-16791: 
Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K1179W mutant
Method: single particle / : Nganga PN, Roderer D, Belyy A, Prumbaum D, Raunser S

EMDB-16792: 
Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K567W K2008W mutant
Method: single particle / : Nganga PN, Roderer D, Belyy A, Prumbaum D, Raunser S

EMDB-16793: 
Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, C16S, C20S, C870S, T1279C mutant
Method: single particle / : Nganga PN, Roderer D, Belyy A, Prumbaum D, Raunser S

EMDB-34929: 
Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form
Method: single particle / : Xu Y, Wu Y, Zhang Y, Fan R, Yang Y, Li D, Yang B, Zhang Z, Dong C, Zhang X, Tang X, Dong H

EMDB-34927: 
Cryo-EM structure of monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex without DNA at 2.76 angostram
Method: single particle / : Xu Y, Wu Y, Zhang Y, Fan R, Yang Y, Li D, Yang B, Zhang Z, Dong C, Zhang X, Tang X, Dong H
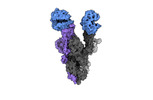
EMDB-40976: 
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to two mink ACE2 receptors
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Zhou B, Liang B

EMDB-40977: 
Cryo-EM structure of mink variant Y453F trimeric spike protein
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Zhou B, Liang B
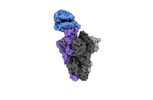
EMDB-40978: 
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors at downRBD conformation
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Zhou B, Liang B

EMDB-40979: 
Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at upRBD conformation
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Zhou B, Liang B

EMDB-40980: 
Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at downRBD conformation.
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Zhou B, Liang B
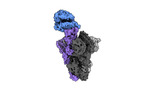
EMDB-41143: 
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Liang B

PDB-8t20: 
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to two mink ACE2 receptors
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Zhou B, Liang B

PDB-8t21: 
Cryo-EM structure of mink variant Y453F trimeric spike protein
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Zhou B, Liang B

PDB-8t22: 
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors at downRBD conformation
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Zhou B, Liang B

PDB-8t23: 
Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at upRBD conformation
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Zhou B, Liang B

PDB-8t25: 
Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at downRBD conformation.
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Zhou B, Liang B

PDB-8taz: 
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Liang B

EMDB-34259: 
cryo-EM structure of Omicron BA.5 S protein in complex with XGv282
Method: single particle / : Xia XY, Zhang YY, Chi XM, Huang BD, Wu LS, Zhou Q

EMDB-34260: 
Cryo-EM map of Omicron BA.5 S protein in complex with XGv282 focused on RBD_XGv282 sub-complex
Method: single particle / : Xia LY, Zhang YY, Zhou Q
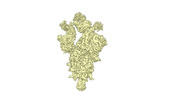
EMDB-34261: 
cryo-EM structure of Omicron BA.5 S protein in complex with XGv289
Method: single particle / : Xia XY, Zhang YY, Chi XM, Huang BD, Wu LS, Zhou Q

EMDB-34262: 
Cryo-EM map of Omicron BA.5 S protein in complex with XGv289 focused on RBD_XGv289 sub-complex
Method: single particle / : Xia LY, Zhang YY, Zhou Q
EMDB-34263: 
cryo-EM structure of Omicron BA.5 S protein in complex with S2L20
Method: single particle / : Xia XY, Zhang YY, Chi XM, Huang BD, Wu LS, Zhou Q

EMDB-34264: 
Cryo-EM map of Omicron BA.5 S protein in complex with S2L20 focused on NTD_S2L20 sub-complex
Method: single particle / : Xia LY, Zhang YY, Zhou Q

EMDB-33489: 
Cryo-EM structure of human DICER-pre-miRNA in a dicing state
Method: single particle / : Lee H, Roh SH

EMDB-33490: 
Cryo-EM structure of an apo-form of human DICER
Method: single particle / : Lee H, Roh SH

EMDB-30947: 
Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state
Method: single particle / : Guo YY, Zhang YY, Yan RH, Huang BD, Ye FF, Wu LS, Chi XM, Zhou Q

EMDB-30948: 
Cryo EM structure of a K+-bound Na+,K+-ATPase in the E2 state
Method: single particle / : Guo YY, Zhang YY, Yan RH, Huang BD, Ye FF, Wu LS, Chi XM, Zhou Q

EMDB-30949: 
Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state with ATP-gamma-S
Method: single particle / : Guo YY, Zhang YY, Yan RH, Huang BD, Ye FF, Wu LS, Chi XM, Zhou Q

EMDB-32869: 
S protein of Delta variant in complex with ZWD12
Method: single particle / : Guo YY, Zhang YY, Zhou Q

EMDB-32870: 
S protein of Delta variant in complex with ZWD12 focused on RBD_ZWD12 sub-complex
Method: single particle / : Guo YY, Zhang YY

EMDB-32871: 
S protein of Delta variant in complex with ZWC6
Method: single particle / : Guo YY, Zhang YY, Zhou Q

EMDB-32872: 
S protein of Delta variant in complex with ZWC6 focused on RBD_ZWC6 sub-complex
Method: single particle / : Guo YY, Zhang YY

EMDB-12566: 
Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B
Method: single particle / : Baronina A, Pike ACW, Yu X, Dong YY, Shintre CA, Tessitore A, Chu A, Rotty B, Venkaya S, Mukhopadhyay SMM, Borkowska O, Chalk R, Shrestha L, Burgess-Brown NA, Edwards AM, Arrowsmith CH, Bountra C, Han S, Carpenter EP, Structural Genomics Consortium (SGC)

EMDB-32920: 
S protein of SARS-CoV-2 in complex with 2G1
Method: single particle / : Guo YY, Zhang YY

EMDB-32921: 
S protein of SARS-CoV-2 in complex with 2G1 focused on RBD_2G1 sub-complex
Method: single particle / : Guo YY, Zhang YY

EMDB-30650: 
Cryo-EM structure of the Ams1 and Nbr1 complex
Method: single particle / : Zhang J, Ye K

EMDB-30652: 
Cryo-EM structure of the Ape4 and Nbr1 complex
Method: single particle / : Zhang J, Ye K

EMDB-30622: 
Cryo-EM structure of amyloid fibril formed by human RIPK3
Method: helical / : Zhao K, Ma YY

EMDB-10094: 
Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with TFB-TBOA
Method: single particle / : Baronina A, Pike ACW, Yu X, Dong YY, Shintre CA, Tessitore A, Chu A, Rotty B, Venkaya S, Mukhopadhyay S, Borkowska O, Chalk R, Shrestha L, Burgess-Brown NA, Edwards AM, Arrowsmith CH, Bountra C, Han S, Carpenter EP, Structural Genomics Consortium (SGC)

EMDB-30276: 
cryo EM map of the S protein of SARS-CoV-2 in complex bound with 4A8
Method: single particle / : Yan RH, Zhang YY, Guo YY, Li YN, Xia L, Zhou Q

EMDB-30277: 
Cryo EM map of the interface between NTD of SARS-CoV-2 and 4A8
Method: single particle / : Yan RH, Zhang YY, Guo YY, Li YN, Xia L, Zhou Q

EMDB-9187: 
Cryo-EM structure of the human neutral amino acid transporter ASCT2
Method: single particle / : Yu X, Han S

EMDB-9188: 
Cryo-EM structure of the human neutral amino acid transporter ASCT2
Method: single particle / : Yu X, Han S
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
